Computational gene regulatory network inference method
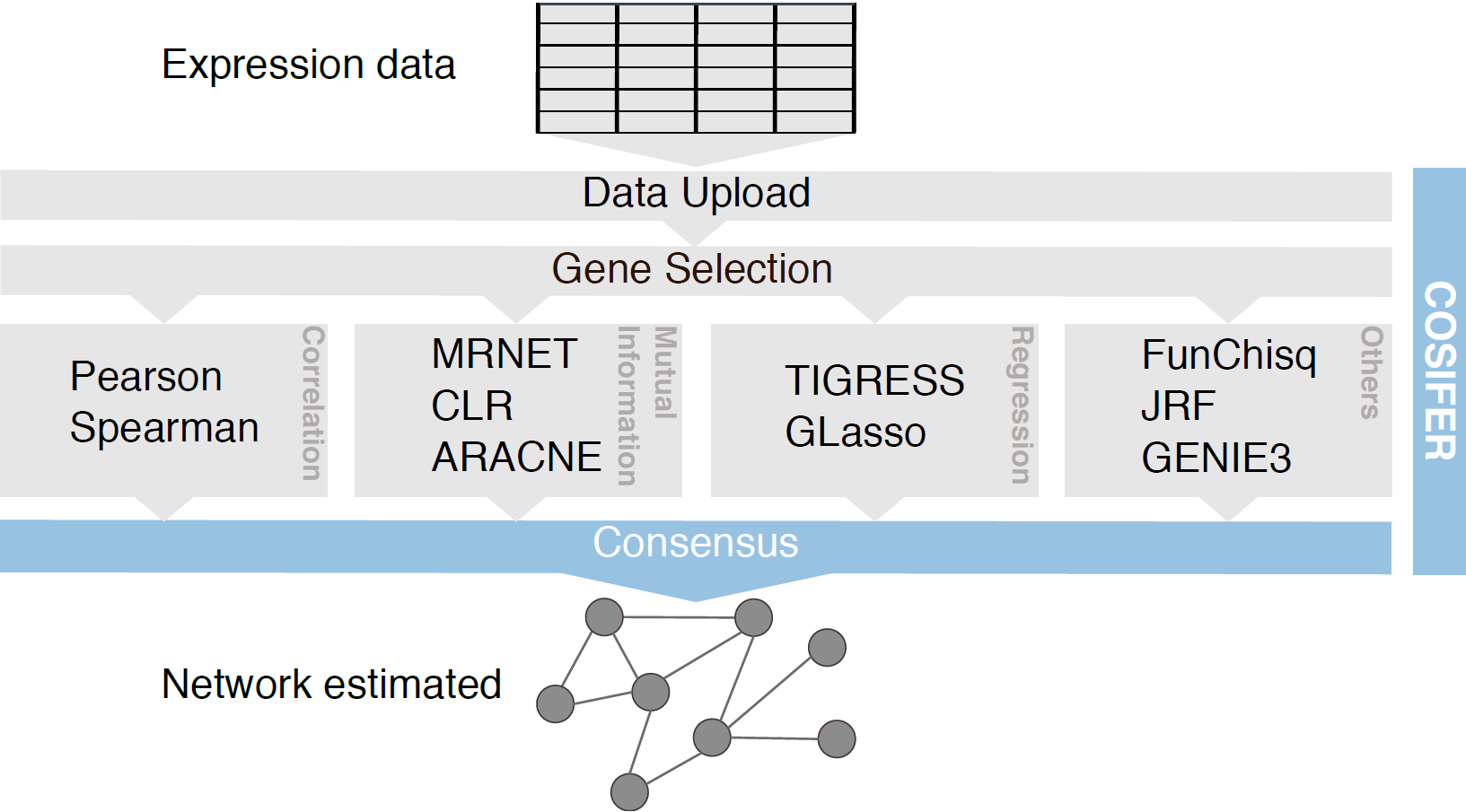
Intracellular networks regulate every kind of cellular decision such as differentiation, proliferation and apoptosis. When these control mechanisms fail, cancer and other diseases may occur. The complexity of these networks originates from the large number and various interactions of the molecules involved. High-throughput technologies such as microarrays and RNA sequencing provide snapshots of the transcriptome and enable insights into the internal regulatory apparatus of a cell. However, inferring the topology of these networks and identifying their key regulators is a challenging task, and international consortia have intensively worked on the development of computational methods tackling this problem. Despite the efforts to compare and develop gene regulatory network inference methods, the research community still lacks easy to access inference tools available to everyone. COSIFER is a web-based platform providing a service for the inference of molecular networks using a consensus between state-of-the-art methodologies given molecular measurements and a list of molecular entities of interest.
Questions?
Ask the expert

Matteo Manica
IBM Research scientist
Funding
This research is funded by the PrECISE EU project
