In histopathology, pathologists assess patient biopsies and tissue resections to study the presence and/or grade of a disease, but also for selecting personalized treatment and monitoring. With respect to other diagnostic technologies, tissue analysis is more invasive, but at the same time provides much higher resolution.
Currently, pathologists assess tissues under a microscope, leading to diagnoses affected by subjective judgment and intra- and inter-observer variability. This is due to the difficulty of the process. The staining intensity in the tissue, as well as the morphological and cellular architecture indicate cancer and many diseases.
“We aim to develop image analytics that can quantify pathogenesis in a high-throughput, bias-free and robust way. ”
—Maria Gabrani, IBM scientist
However, disease susceptibility and progression is a complex, multifactorial molecular process. Diseases such as cancer exhibit tissue and cellular heterogeneities, which impedes differentiation between different stages or types of cell formations. At the same time, the procedure is time-consuming and low-throughput, strained by the number of tissue samples generated per day.
Emerging initiatives to bring hospitals into the digital era are using bright-field and fluorescence scanners to convert glass slides of tissue specimens and needle biopsies into virtual microscopy images of very high quality, enabling digital image analysis.
Our focus
At IBM Research – Zurich, we are focusing on the analysis of digitized histopathology and molecular expression images, as well as cytology images. Imaging tissue specimens is a powerful tool to extract quantitative metrics of phenotypic properties while preserving the morphology and spatial relationship of the tissue microenvironment.
Novel staining technologies such as immunohistochemistry (IHC) and in situ hybridization (ISH) further improve the evidencing of molecular expression patterns by means of multicolor visualization.
Such techniques are thus commonly used for predicting disease susceptibility and stratification as well as for selecting treatment and monitoring. However, translating molecular expression imaging into direct health benefits has been slow.
Two major factors contribute to that. On the one hand, disease susceptibility and progression is a complex, multifactorial molecular process. The tissue and cell heterogeneity exhibited by diseases such as cancer occur most prominently between inflammatory response and malignant cell transition.
On the other hand, the relative quantification of the selected features in stained tissue samples is ambiguous, tedious and time-consuming, and therefore prone to technician and clerical errors. This in turn leads to intra- and interobserver variability and low throughput.
At IBM Research – Zurich, we are developing advanced image analytics to address both the above limitations. Our aim is to transform the analysis of stained tissue images into a high-throughput, robust, quantitative and data-driven science.
Value proposition
Patient
- Higher accuracy
- Fewer misdiagnoses
- Better outcomes
Pathologist
- Case ordering
- Efficiency
- Surgery assistance
Hospital
- Case ordering
- Less diagnostic variability
- Lower costs
Payer
- Second-opinion enablement
- Lower costs
Complexities

Diagnostic complexity
- Misdiagnosis
- Over-diagnosis
- Over-treatment
Phenotypic complexity
- Tissue/tumor/cell/molecular heterogeneity
- Cell plasticity

Process complexity
- Capturing induced ambiguity
- Time consuming
- Tedious
- Bias prone

Disease complexity
- Molecular
- Cellular/Tissue
- Historical
- Environmental
Projects
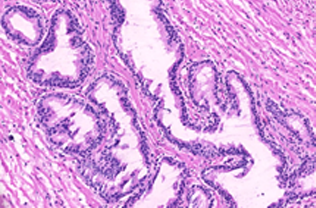
Prostate cancer stratification
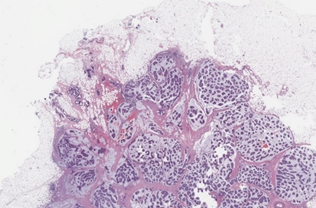
Breast cancer tumor proliferation estimation
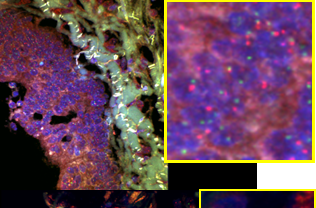
Colorectal cancer metastasis study
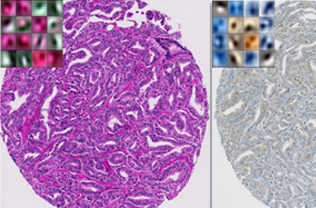
Tissue heterogeneity quantification
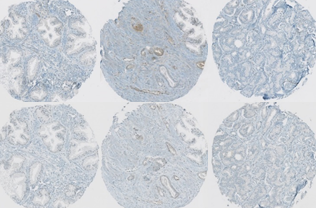
Protein signature quantification
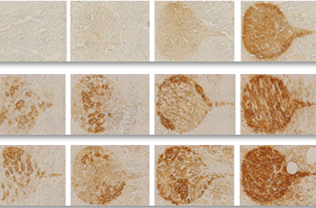
Stained image quality metric and sensitivity to staining parameters estimation
Interviews & press articles
- Potential for the use of artificial intelligence in the health sector, Reuters
- Künstliche Intelligenz – Die Macht der Maschinen, SRF Einstein
- Comment le big data propulse la médecine dans une nouvelle dimension, RTS
- Forscher bringen dem Computer “Watson” bei, Brustkrebs auf Bildern zu erkennen, was er inzwischen besser als Ärzte kann, 3sat Nano

Collaboration
We base our technology development and validation on data, labels and domain insights from experts in the field through numerous collaborations with university hospitals and other partners, such as the University Hospital Zurich, Switzerland, and Humanitas University Hospital, Italy, as they are nicely described in the TV interviews.
Journals
- T. Binder, et al.,
“Multi-organ pathology image segmentation using deep learning,”
Frontiers in Medicine–Pathology 6, 173, 2019. - A. Anghel et al.,
“A High-Performance System for Robust Stain Normalization of Whole-Slide Images in Histopathology,”
Frontiers in Medicine 6, 193, 2019. - A. Kashyap et al.,
“Quantitative microimmunohistochemistry (qμIC): a method to grade immunostains in tumor tissues using saturation kinetics,”
Nature Biomedical Engineering 3(6) 478–490, 2019. - N.M. Arar et al.,
“High-Quality Immunohistochemical Stains through Computational Assay Parameter Optimization,”
IEEE Transactions on Biomedical Engineering, 2019.
- M. Veta et al.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge,
Medical Image Analysis 54, 111-121, 2018. - E. Zerhouni, et al.,
“Computational Processing of Histological Images,”
ERCIM News 108, 14-15, 2017. - Q. Zhong, et al.,
“Image-based computational quantification and visualization of genetic alterations and tumour heterogeneity,”
Scientic Reports 6(24146), 2016. - E. Zerhouni, et al.,
“Big Data Takes on Prostate Cancer,”
ERCIM News 104, 34-35, 2016.
Conferences
- G. Jaumeet al.,
“edGNN: A simple and powerful GNN for directed labeled graphs,”
ICLR 2019, workshop on graphs and manifolds. - N. Ioannou et al.,
“Accelerated ML-assisted Tumor Detection in High-Resolution Histopathology Images,”
MICCAI, LNCS, Vol. 11764, 406–414, 2019. - P. Pati, et al.,
“A deep learning framework for context-aware mitotic activity estimation in whole slide images,”
SPIE Medical Imaging, Vol. 10956, 2019. - M. Stanisavljevic, et al.,
“A Fast and Scalable Pipeline for Stain Normalization of Whole-Slide Images in Histopathology,”
In Computer Vision: Proc. ECCV Bioimage Computing Workshops, LNCS, Springer, 2018. - Pushpak Pati et al.
“Region-of-Interest Localization in Whole-Slide Images: A Semi-Supervised Approach for Positive Unlabeled Data,”
Proc. SPIE Medical Imaging: Digital Pathology 10581, 2018.
- N.M. Arar, et al.,
“Computational Immunohistochemistry: Recipes for Standardization of Immunostaining,”
Proc. International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), 48–55, 2017. - E. Zerhouni, et al.,
“Heterogeneity Characterization of Immunohistochemically Stained Tissue using Convolutional Autoencoder,”
Proc. SPIE 10140, Medical Imaging 2017: Digital Pathology, 101400P, 2017. - E. Zerhouni, et al.,
“Wide Residual Networks for Mitosis Detection,”
Proc. IEEE 14th International Symposium on Biomedical Imaging (ISBI), 2017. - E. Zerhouni, et al.,
“Disease Grading of Heterogeneous Tissue using Convolutional Autoencoder,”
Proc. IEEE 14th International Symposium on Biomedical Imaging (ISBI), 2017. - E. Zerhouni, et al.,
“Deciphering protein signatures using color, morphological, and topological analysis of immunohistochemically stained human tissues,”
Proc. SPIE, Vol. 9791, 97910T, 2016. - E. Zerhouni, et al.,
“A computational framework for disease grading using protein signatures,”
Proc. 13th IEEE International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 1401–1404, 2016.
Presentations
- P. Pati, et al.,
“Mitotic cell density estimation on whole tumor region based on deep learning,”
DGP 2018, Berlin, Germany, May 24-26, 2018. - E. Zerhouni, et al.,
“A Computationaly extracting unique protein signatures for IHC assays per grade and protein for personalized treatment,”
DGP 2016, Berlin, Germany, May 19-21, 2016. - M. Kolectou, et al.,
“A Computational framework for systems pathology of prostate cancer,”
DGP 2016, Berlin, Germany, May 19-21, 2016. - M. Kolectou, et al.,
“A Computational framework for systems pathology of prostate cancer,”
Systems Biology of Human Diseases, Boston, MA, USA, June 14-16, 2016. - Q. Zhong, et al.,
“Computational profiling of heterogeneity reveals high concordance between morphology- and proteomics based methods in prostate cancer,”
DGP 2015, Frankfurt, Germany, May 28-31, 2015.
