Our mission
The Medical Decision Support group is all about providing support to all stakeholders in the healthcare system to accelerate processes and shorten time to treatment.
Patients
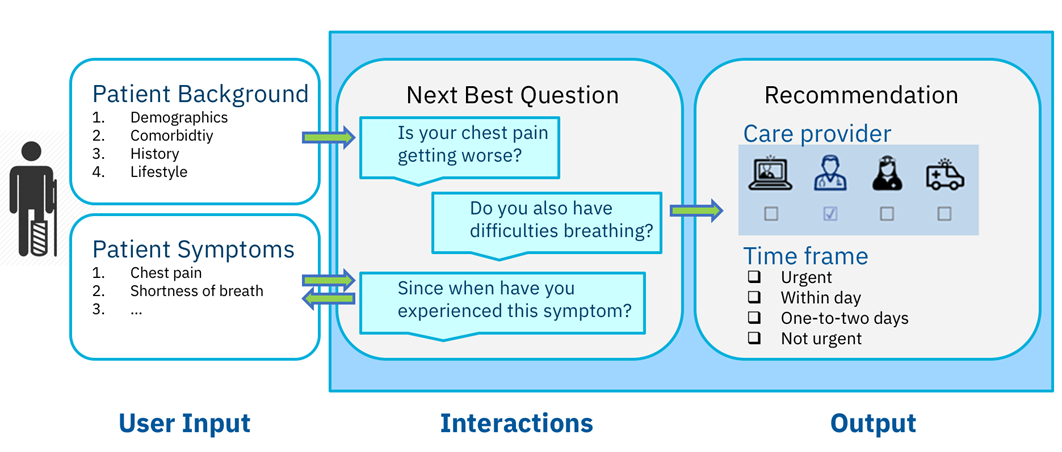
Our medical triage solution guides patients to the appropriate healthcare provider in a timely manner instead of simply checking for symptoms and making a possible diagnosis.
Caregivers
Our work on rare diseases supports medical staff in order to accelerate the differential diagnosis process.
Administrators
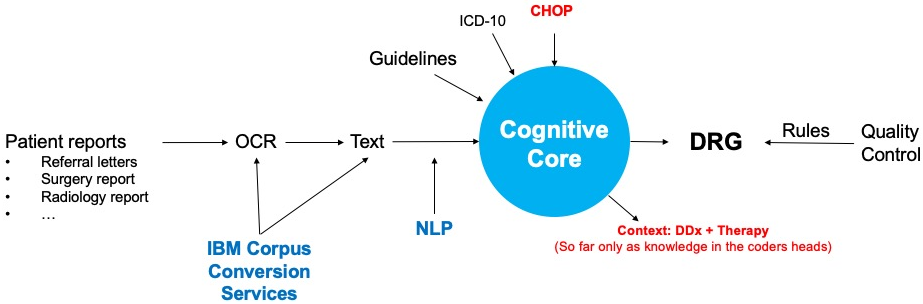
Facilitating medical coding addresses major pain points of administrative staff by allowing them to focus on higher-quality tasks and transforming the invoicing of medical services between hospitals and insurances from a manual, cumbersome chore into an automated process with human supervision.
Process acceleration in healthcare
Solution 1: Cognitive medical triage
Decision support in healthcare is a major source of process optimization in the healthcare system. Decisions for stakeholders can be facilitated using cognitive technology.
Our cognitive medical triage solution is not yet another symptom checker for patients. The interactive system requests information about the patient’s health status, ant then guides the patient to the appropriate care provider to obtain suitable medical care and support in a timely manner. This is a major differentiator because patients require the knowledge to seek medical help, not merely a diagnosis. The solution is so advanced that it is currently undergoing medical certification and production, and is imminently scheduled to be launched with a telemedicine provider.
Solution 2: Cognitive medical coding
Medical coding is currently a manual process. However, there is a lack of human resources with the required highly specialized skill set available on the market required. Our solution, which is based on natural language processing and knowledge graph technology, solves these issues by automating parts of the process. It augments the coder’s work, who can now focus on higher-quality tasks. Furthermore, it changes the healthcare ecosystem in the sense that invoicing will become an automatic transaction rather than a long manual process involving multiple parties. In this way, our solution contributes to creating resilient and sustainable healthcare systems.
Ask the experts

Adam Ivankay
PhD student
Publications
[1] Girardi I. et al.
“Patient Risk Assessment and Warning Symptom Detection Using Deep Attention-Based Neural Networks”
Proc. 9th Int’l Workshop on Health Text Mining and Information Analysis
(EMNLP 2018), Association for Computational Linguistics, Brussels, Belgium, 2018.
[2] Girardi I. et al.
“Von vertraulicher Forschung zur radikalen Kollaboration”
in Zukunftsmanagement in Gesundheitssystemen, Rebscher H., Kaufmann S. (eds.) ISBN 978-3-86216-482-0, Medhochzwei, 87–100, 2018.
[3] Auer C. et al.
“Künstliche Intelligenz im Gesundheitswesen”
Themenband Digitale Gesundheit, Springer, 2018.
[4] Reumann M. et al.
“Cognitive DDx Assistant in Rare Diseases”
Proc. IEEE Conf. Eng. Med. Biol. Soc., 3244–3247, 2018.
[5] Mählmann L. et al.
“Big Data for Public Health Policy Making: Policy Empowerment”
Public Health Genomics 20(6), 312–320, 2018.
[6] Van Roessel I. et al.
“Potentials and challenges of the health data cooperative model”
Public Health Genomics 20(6), 321–331, 2018.
[7] Hollenstein N. et al.
“An Ontology for the Swiss Operation Classification System (CHOP)”
Swiss Medical Informatics (accepted for publication), 2018.
[8] Reumann M.
“Big Data in Public Health: From Genes to Society”
Universitaire Pers Maastricht. ISBN 978-94-6159-753-3, 2017.
[9] Evangelatos N. et al.
“Metabolomics in Sepsis and Its Impact on Public Health”
Public Health Genomics 20(5), 274–285, 2017.
[10] Schee genannt Halfmann S. et al.
“Personalized Medicine: What’s in it for Rare Diseases?”
Advances in Experimental Medicine and Biology 1031, 387–404, 2017.
[11] Reumann M. et al.
“Big Data und kognitive Assistenzsysteme verändern die Versorgung”
in Digitalisierungsmanagement in Gesundheitssystemen, Rebscher H., Kaufmann S. (eds.), ISBN 978-3-86216-367-0, Medhochzwei, 99–118, 2017.
[12] Reumann M., Böttcher B.
“Der lernende Assistent in der digitalen Revolution”
Health Care Management 4, 28–29, 2017.
[13] Leyens L. et al.
“Use of Big Data for drug development and for Public and Personal Health and Care”
Genetic Epidemiology 41(1), 51–60, 2017.
[14] Reumann M., Böttcher B.
“Cognitive Computing: Essenzieller Teil der Medizin 2020”
TumorDiagnostik & Therapie 37(6), 322–326, 2016.
[15] Evangelatos N. et al.
“Clinical Trial Data as Public Goods: Fair Trade and the Virtual Knowledge Bank as a Solution to the Free Rider Problem — A Framework for the Promotion of Innovation by Facilitation of Clinical Trial Data Sharing among Biopharmaceutical Companies in the Era of Omics and Big Data”
Public Health Genomics 19, 211–219, 2016.
[16] MacInnis R.J. et al.
“Use of a novel non-parametric version of DEPTH to identify genomic regions associated with prostate cancer risk”
Cancer Epidemiol Biomarkers Prev. 25(12), 1619–1624, 2016.
[17] Reumann M., Böttcher B.
“Cognitive Computing: Essenzieller Teil der Medizin 2020”
Klinikarzt 45(5), 238–242, 2016.
[18] Reumann M.
“Big data and cognitive computing in healthcare: Weathering the perfect storm”
IEEE MELECON, Limassol, Cyprus, Extended Abstract, 2016.
[19] Reumann M.
“Innovationshürden für die digitale Gesundheit — eine persönliche Sicht”
Stiftung Münch Themen, 2016.
[20] Studer L. et al.
“Marginalized continuous time Bayesian networks for network reconstruction from incomplete observations”
AAAI Conference on Artificial Intelligence (AAAI ’16) Phoenix, AZ, USA, 2016.
[21] Reumann M.
“Big Data: Übernehmen Superrechner wie Watson das Kommando in der Medizin?”
AGAPLESION Wissen, 2015.
[22] Reumann M.
“Translating Innovative Supercomputing into Clinical Application: Challenges and Opportunities”
CLINAM, Basel, Switzerland (Extended Abstract), 2015.
[23] Reumann M.
“Big Data and cognitive computing in life sciences: Today’s challenges and a bright future”
Proc. IEEE Eng. Med. Biol. Soc., 2015.
[24] Reumann M.
“Big Data in life science: Harnessing supercomputing to make Big Data accessible and actionable”
PASC’15. Zürich, (Abstract), 2015.
[25] Reumann M.
“Übernehmen Supercomputer das Kommando in der Medizin?”
f&w Führen und Wirtschaften im Krankenhaus 4, 228–230, 2015.
[26] Goudey B.W. et al.
“High performance computing enabling exhaustive analysis of higher order single nucleotide polymorphism interaction in genome wide association studies”
Health Information Science and Systems 3 (Suppl. 1), S3, 2015.
[27] Rusu L. et al.
“Harnessing process pipelines for leveraging next generation sequencing in microbiological diagnostics”
Health Information Science and Systems 3 (Suppl 1), S7, 2015.
[28] Rogers P. et al.
“Informatics for enabling enterprise-scale genomics”
53rd ASMR National Scientific Conference on Transdisciplinary Research (Abstract), 2014.
[29] Reumann M.
“Big Data and cognitive computing in life sciences: today’s challenges and a bright future”
Nürnberger Kreis “The internet of things and people — wie die Digitalisierung unsere Welt verändert” (Abstract), 2014.
[30] Reumann M.
“Model driven cognitive computing and analytics in systems medicine”
IKMB Summer School on Systems Medicine, Kiel, Germany (Abstract), 2014.
[31] Davies M. et al.
“Intuitive information and knowledge representation of disease incidence and respective intervention strategies”
Studies in Health Technology and Informatics 205, 1173–1177, 2014.
[32] Wyres K.L. et al.
“WGS Analysis and Interpretation in Clinical and Public Health Microbiology Laboratories: What are the Requirements and how do Existing Tools Compare?”
Pathogens. 3(2), 437–458, 2014.
[33] Reumann M.
“Multiscale Systems Biology: Big Data challenges in supercomputing enabling translational medicine in cardiology”
International Supercomputing Conference ISC’14, Leipzig, Germany (Abstract), 2014.
[34] Davis M. et al.
“Spatio-temporal information and knowledge representation of disease incidence and respective intervention strategies”
Stud. Health Technol. Inform. 205, 1173–1177, 2014.
[35] Mirmomeni M. et al.
“Resolving ambiguity in genome assembly using high performance computing”
HISA Big Data 2014 Conference, Melbourne, Australia, 2014.
[36] Mirmomeni M. et al.
“Resolving ambiguity in genome assembly using high performance computing”
IBM Research Report RC25439 (MRL1401–001), 2014.
[37] Mehnhorn F, Reumann M.
“Genome assembly framework on massively parallel, distributed memory supercomputers”
Biomed. Tech. 58 (Suppl. 1), 2013.
[38] Richards D.F. et al.
“Toward Real-time Simulation of Cardiac Electrophysiology in a Human Heart at Near-Cellular Resolution”
Comp Methods in Biomech and Biomed Eng. 6(7), 802–805, 2013.
[39] Reumann M. et al.
“Big Data challenges in predictive modeling for treatment using biophysical cardiac models”
HISA Big Data Conference, Melbourne, VIC, Australia, 2013.
[40] Goudey B.W. et al.
“High performance computing enabling exhaustive analysis of higher order single nucleotide polymorphism interaction in Genome Wide Association Studies”
HISA Big Data Conference, Melbourne, VIC, Australia, 2013.
[41] Rusu L. et al.
“Harnessing process pipelines for leveraging next generation sequencing in microbiological diagnostics”
HISA Big Data Conference, Melbourne, VIC, Australia, 2013.
[42] Garg S. et al.
“Stream computing in a children’s intensive care unit: enabling investigation of age dependent predictors for pro-active intervention”
HISA Big Data Conference, Melbourne, VIC, Australia, 2013.
[43] Rao C. et al.
“The IBM Watson solution for Healthcare bringing knowledge based analytics into clinical practice”
HISA Big Data Conference, Melbourne, VIC, Australia, 2013.
[44] Garnavi R., Reumann M., Sun X.
“IBM Patient Care and Insights delivering an integrated solution for advanced analytics in healthcare”
HISA Big Data Conference, Melbourne, VIC, Australia, 2013.
[45] Sun X. et al.
“Facilitating Clinical Data Analytics for Comparative Effectiveness Research”
HISA Big Data Conference, Melbourne, VIC, Australia, 2013.
[46] Sun X. et al.
“Facilitating Clinical Data Analytics for Comparative Effectiveness Research”
IBM Research Report RC25357 (AUS1303–001), 2013.
[47] Rice J.J. et al.
“Highly scalable cardiac modeling codes for petascale computing”
SIAM Computational Science and Engineering Meeting in Boston, 2013.
[48] Carnes B. et al.
“Science at LLNL with IBM Blue Gene/Q”
IBM Journal of Research and Development 57(1/2), 11:1–11:18,
IF: 4.863, 2013.
[49] Reumann, M. et al.
“Supercomputing enabling exhaustive statistical analysis of genome wide association study data: Preliminary results”
Proc. Annual International Conf.
of the IEEE Engineering in Medicine and Biology Society, 2012.